GEMMER
- Complements the existing web-based databases and visualization tools for budding yeast
- Provides high-quality visualizations of interaction networks for user-specified gene(s)
- Highlights within such visualizations information on function, expression, timing and network importance
- Goes beyond "hairball" networks to provide rational visualizations
Data integration
GEMMER integrates data from various sources into a single database
- SGD Interactome and experimental evidence
- CYCLoPs Relative abundance and localization
- YeastGFP Absolute abundance and localization
- SCEPTRANS Timing of peak transcription during the cell cycle
- Yeast 7.6 List of metabolic enzymes
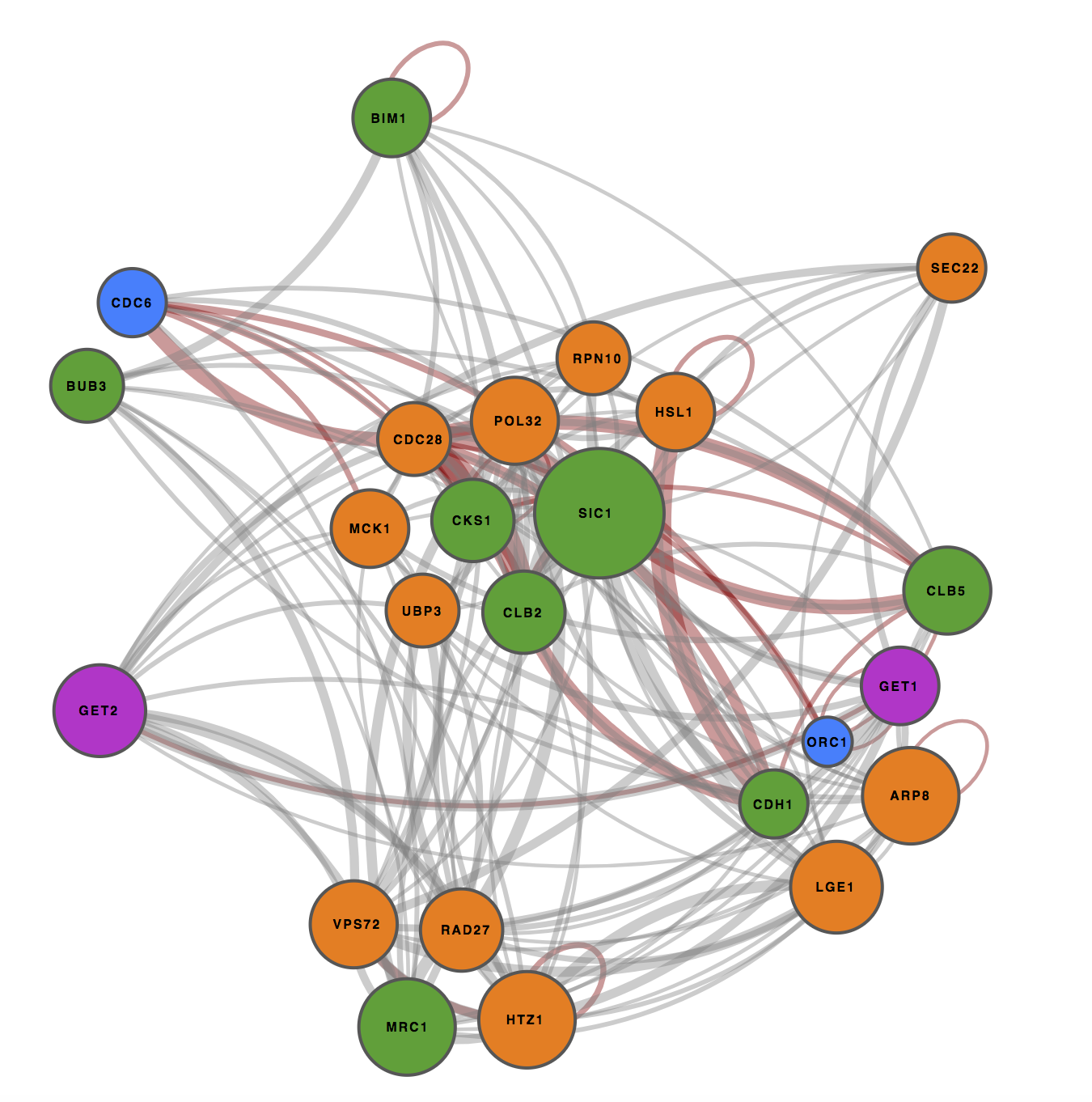
Network visualizations
User queries result in interactomes that may be visualized with various layoutsSee the examples for more inspiration
Data export and modeling
Visualized interactomes may be exported in a variety of formats for further analysisExport network to Excel, JSON, GraphML
For offline usage and further analysis by Gephi and Cytoscape